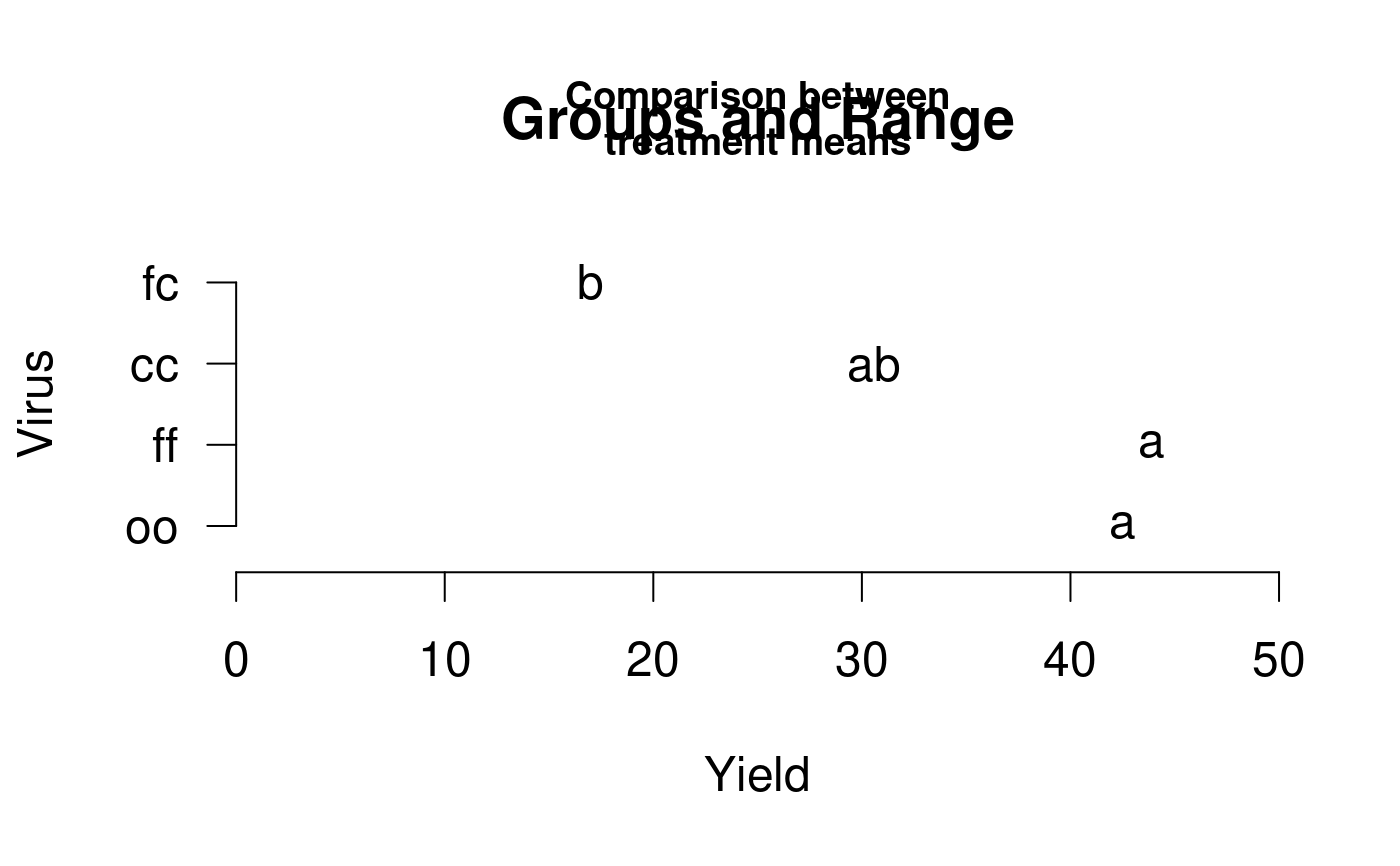
It plots bars of the averages of treatments to compare. It uses the objects generated by a procedure of comparison like LSD, HSD, Kruskall, Waller-Duncan, Friedman or Durbin. It can also display the 'average' value over each bar in a bar chart.
# S3 method for group plot( x, variation = c("range", "IQR", "SE", "SD"), horiz = FALSE, col = NULL, xlim = NULL, ylim = NULL, main = NULL, cex = NULL, hy = 0, ... )
Arguments
| x | Object created by a test of comparison |
|---|---|
| variation | in lines by range, IQR, standard deviation or error |
| horiz | Horizontal or vertical image |
| col | line colors |
| xlim | optional, axis x limits |
| ylim | optional, axis y limits |
| main | optional, main title |
| cex | optional, group label size |
| hy | optional, default =0, sum group label position |
| ... | Parameters of the function barplot() |
Details
The output is a vector that indicates the position of the treatments on the coordinate axes.
See also
BIB.test, DAU.test,
duncan.test, durbin.test,
friedman, HSD.test, kruskal,
LSD.test, Median.test, PBIB.test,
REGW.test, scheffe.test, SNK.test,
waerden.test, waller.test
Examples
library(agricolae) data(sweetpotato) model<-aov(yield~virus,data=sweetpotato) comparison<- LSD.test(model,"virus",alpha=0.01,group=TRUE) #startgraph par(cex=1.5) plot(comparison,horiz=TRUE,xlim=c(0,50),las=1)#> Warning: NAs introduced by coercion#endgraph