This test is adapted from the Newman-Keuls method. Duncan's test does not control family wise error rate at the specified alpha level. It has more power than the other post tests, but only because it doesn't control the error rate properly. The Experimentwise Error Rate at: 1-(1-alpha)^(a-1); where "a" is the number of means and is the Per-Comparison Error Rate. Duncan's procedure is only very slightly more conservative than LSD. The level by alpha default is 0.05.
duncan.test( y, trt, DFerror, MSerror, alpha = 0.05, group = TRUE, main = NULL, console = FALSE )
Arguments
| y | model(aov or lm) or answer of the experimental unit |
|---|---|
| trt | Constant( only y=model) or vector treatment applied to each experimental unit |
| DFerror | Degree free |
| MSerror | Mean Square Error |
| alpha | Significant level |
| group | TRUE or FALSE |
| main | Title |
| console | logical, print output |
Value
Statistics of the model
Design parameters
Critical Range Table
Statistical summary of the study variable
Comparison between treatments
Formation of treatment groups
Details
It is necessary first makes a analysis of variance.
if y = model, then to apply the instruction:
duncan.test(model, "trt",
alpha = 0.05, group = TRUE, main = NULL, console = FALSE)
where the model
class is aov or lm.
References
1. Principles and procedures of statistics a biometrical
approach Steel & Torry & Dickey. Third Edition 1997
2. Multiple
comparisons theory and methods. Departament of statistics the Ohio State
University. USA, 1996. Jason C. Hsu. Chapman Hall/CRC.
See also
BIB.test, DAU.test,
durbin.test, friedman, HSD.test,
kruskal, LSD.test, Median.test,
PBIB.test, REGW.test,
scheffe.test, SNK.test,
waerden.test, waller.test,
plot.group
Examples
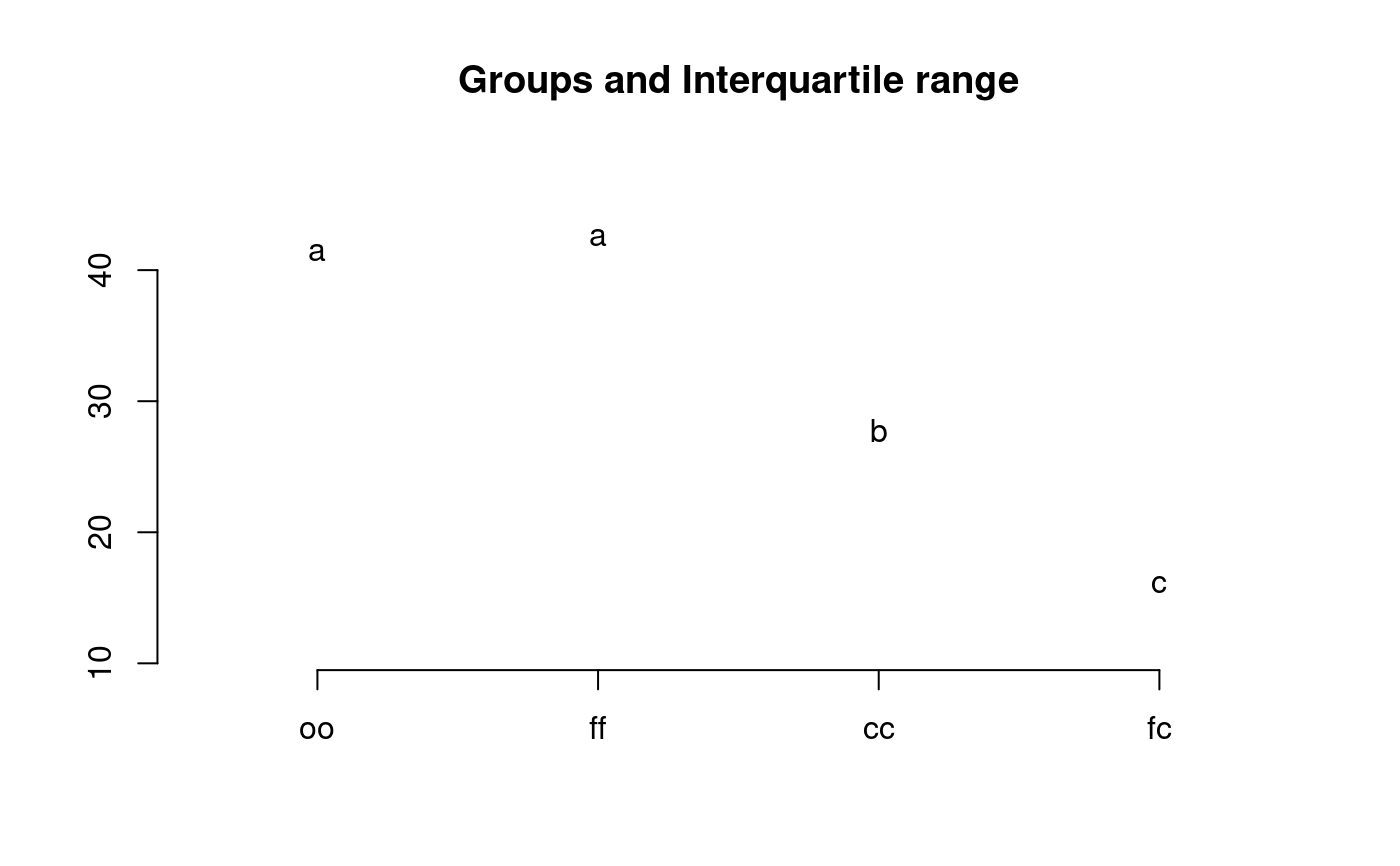
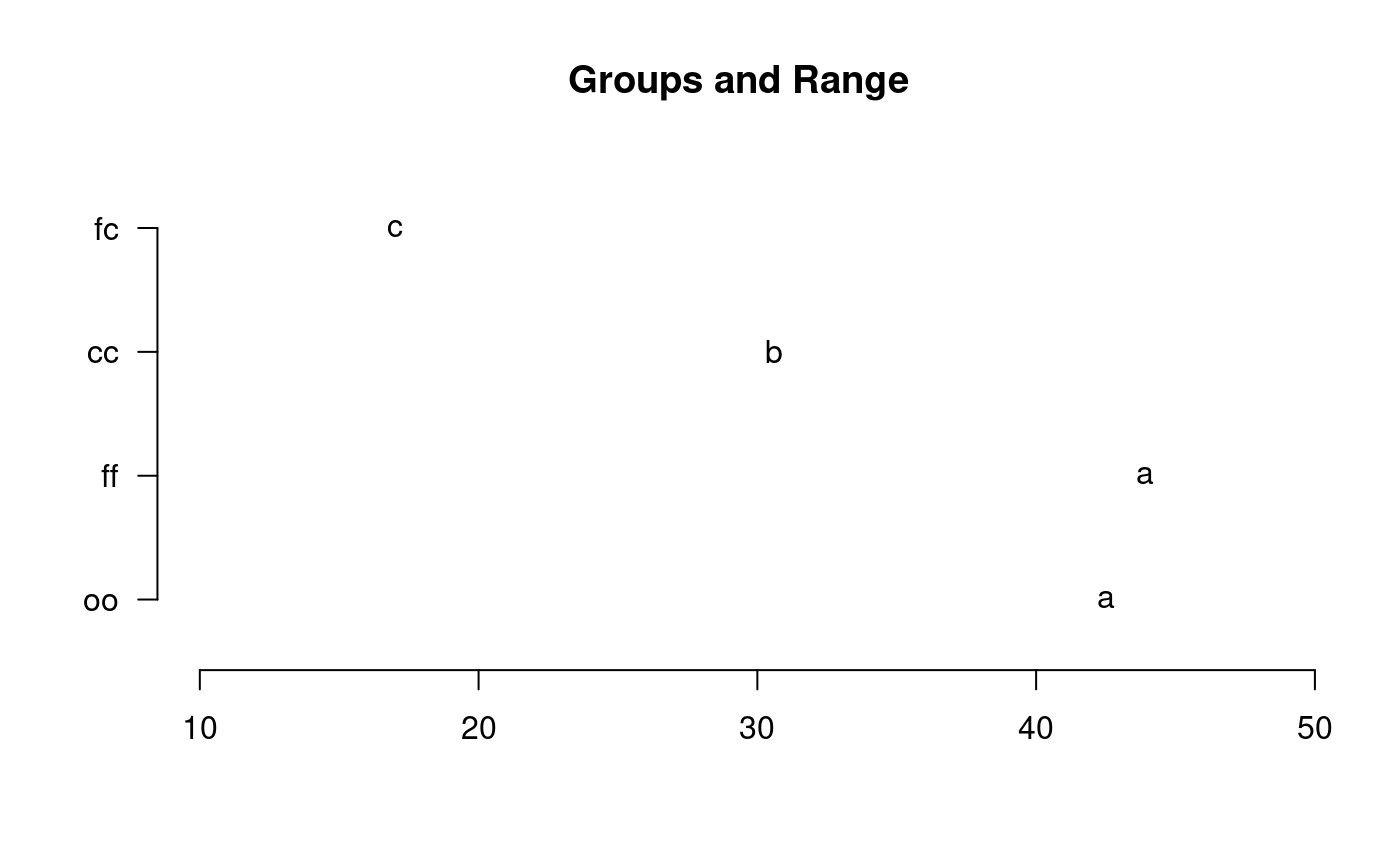
library(agricolae) data(sweetpotato) model<-aov(yield~virus,data=sweetpotato) out <- duncan.test(model,"virus", main="Yield of sweetpotato. Dealt with different virus") plot(out,variation="IQR")#> Warning: NAs introduced by coercionduncan.test(model,"virus",alpha=0.01,console=TRUE)#> #> Study: model ~ "virus" #> #> Duncan's new multiple range test #> for yield #> #> Mean Square Error: 22.48917 #> #> virus, means #> #> yield std r Min Max #> cc 24.40000 3.609709 3 21.7 28.5 #> fc 12.86667 2.159475 3 10.6 14.9 #> ff 36.33333 7.333030 3 28.0 41.8 #> oo 36.90000 4.300000 3 32.1 40.4 #> #> Alpha: 0.01 ; DF Error: 8 #> #> Critical Range #> 2 3 4 #> 12.99223 13.52267 13.84424 #> #> Means with the same letter are not significantly different. #> #> yield groups #> oo 36.90000 a #> ff 36.33333 a #> cc 24.40000 ab #> fc 12.86667 b# version old duncan.test() df<-df.residual(model) MSerror<-deviance(model)/df out <- with(sweetpotato,duncan.test(yield,virus,df,MSerror, group=TRUE)) plot(out,horiz=TRUE,las=1)#> Warning: NAs introduced by coercion#> yield groups #> oo 36.90000 a #> ff 36.33333 a #> cc 24.40000 b #> fc 12.86667 c