The variance analysis of a split-split plot design is divided into three parts: the main-plot, subplot and sub-subplot analysis.
ssp.plot(block, pplot, splot, ssplot, Y)
Arguments
| block | replications |
|---|---|
| pplot | Factor main plot |
| splot | Factor subplot |
| ssplot | Factor sub-subplot |
| Y | Variable, response |
Value
ANOVA: Splip Split plot analysis
Details
The split-split-plot design is an extension of the split-plot design to accommodate a third factor: one factor in main-plot, other in subplot and the third factor in sub-subplot
References
Statistical procedures for agricultural research. Kwanchai A. Gomez, Arturo A. Gomez. Second Edition. 1984.
See also
Examples
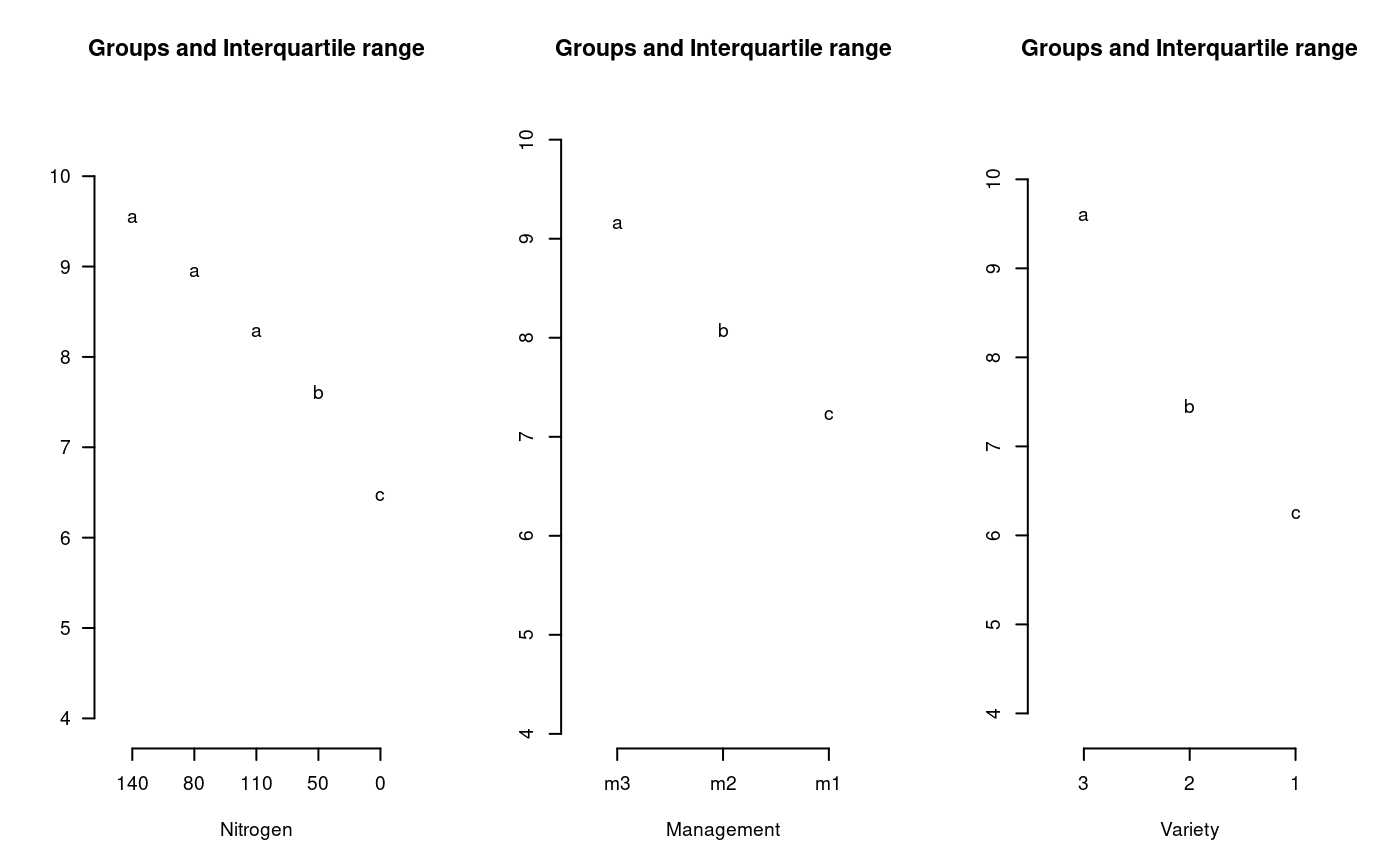
# Statistical procedures for agricultural research, pag 143 # Grain Yields of Three Rice Varieties Grown under #Three Management practices and Five Nitrogen levels; in a #split-split-plot design with nitrogen as main-plot, #management practice as subplot, and variety as sub-subplot #factores, with three replications. library(agricolae) f <- system.file("external/ssp.csv", package="agricolae") ssp<-read.csv(f) model<-with(ssp,ssp.plot(block,nitrogen,management,variety,yield))#> #> ANALYSIS SPLIT-SPLIT PLOT: yield #> Class level information #> #> nitrogen : 0 50 80 110 140 #> management : m1 m2 m3 #> variety : 1 2 3 #> block : 1 2 3 #> #> Number of observations: 135 #> #> Analysis of Variance Table #> #> Response: yield #> Df Sum Sq Mean Sq F value Pr(>F) #> block 2 0.732 0.366 0.6578 0.543910 #> nitrogen 4 61.641 15.410 27.6953 9.734e-05 *** #> Ea 8 4.451 0.556 #> management 2 42.936 21.468 81.9965 2.303e-10 *** #> nitrogen:management 8 1.103 0.138 0.5266 0.822648 #> Eb 20 5.236 0.262 #> variety 2 206.013 103.007 207.8667 < 2.2e-16 *** #> variety:nitrogen 8 14.145 1.768 3.5679 0.001916 ** #> variety:management 4 3.852 0.963 1.9432 0.114899 #> variety:nitrogen:management 16 3.699 0.231 0.4666 0.953759 #> Ec 60 29.732 0.496 #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> cv(a) = 11.4 %, cv(b) = 7.8 %, cv(c) = 10.7 %, Mean = 6.554415 #>gla<-model$gl.a; glb<-model$gl.b; glc<-model$gl.c Ea<-model$Ea; Eb<-model$Eb; Ec<-model$Ec par(mfrow=c(1,3),cex=0.6) out1<-with(ssp,LSD.test(yield,nitrogen,gla,Ea,console=TRUE))#> #> Study: yield ~ nitrogen #> #> LSD t Test for yield #> #> Mean Square Error: 0.5564188 #> #> nitrogen, means and individual ( 95 %) CI #> #> yield std r LCL UCL Min Max #> 0 5.384704 1.206963 27 5.053665 5.715743 3.320 8.020 #> 50 6.220333 1.615861 27 5.889294 6.551372 3.188 9.942 #> 80 6.995741 1.371302 27 6.664702 7.326780 4.422 9.320 #> 110 6.937370 1.474547 27 6.606331 7.268409 4.246 9.660 #> 140 7.233926 1.968153 27 6.902887 7.564965 3.132 10.360 #> #> Alpha: 0.05 ; DF Error: 8 #> Critical Value of t: 2.306004 #> #> least Significant Difference: 0.4681598 #> #> Treatments with the same letter are not significantly different. #> #> yield groups #> 140 7.233926 a #> 80 6.995741 a #> 110 6.937370 a #> 50 6.220333 b #> 0 5.384704 c#> #> Study: yield ~ management #> #> LSD t Test for yield #> #> Mean Square Error: 0.2618167 #> #> management, means and individual ( 95 %) CI #> #> yield std r LCL UCL Min Max #> m1 5.900378 1.486531 45 5.741267 6.059488 3.132 9.314 #> m2 6.486156 1.621936 45 6.327045 6.645266 3.625 9.680 #> m3 7.276711 1.635020 45 7.117601 7.435822 4.225 10.360 #> #> Alpha: 0.05 ; DF Error: 20 #> Critical Value of t: 2.085963 #> #> least Significant Difference: 0.2250164 #> #> Treatments with the same letter are not significantly different. #> #> yield groups #> m3 7.276711 a #> m2 6.486156 b #> m1 5.900378 c#> #> Study: yield ~ variety #> #> LSD t Test for yield #> #> Mean Square Error: 0.4955415 #> #> variety, means and individual ( 95 %) CI #> #> yield std r LCL UCL Min Max #> 1 5.126822 0.971530 45 4.916915 5.336730 3.132 7.309 #> 2 6.396133 1.065752 45 6.186226 6.606041 4.166 8.950 #> 3 8.140289 1.314438 45 7.930381 8.350197 5.244 10.360 #> #> Alpha: 0.05 ; DF Error: 60 #> Critical Value of t: 2.000298 #> #> least Significant Difference: 0.2968543 #> #> Treatments with the same letter are not significantly different. #> #> yield groups #> 3 8.140289 a #> 2 6.396133 b #> 1 5.126822 c#> Warning: NAs introduced by coercion#> Warning: NAs introduced by coercion#> Warning: NAs introduced by coercion# with aov AOV<-aov(yield ~ block + nitrogen*management*variety + Error(block/nitrogen/management),data=ssp) summary(AOV)#> #> Error: block #> Df Sum Sq Mean Sq #> block 1 0.07891 0.07891 #> #> Error: block:nitrogen #> Df Sum Sq Mean Sq #> nitrogen 1 52.13 52.13 #> #> Error: block:nitrogen:management #> Df Sum Sq Mean Sq #> management 2 30.57 15.28 #> #> Error: Within #> Df Sum Sq Mean Sq F value Pr(>F) #> nitrogen 1 4.44 4.44 9.124 0.00309 ** #> management 2 12.93 6.46 13.292 6.16e-06 *** #> variety 1 204.32 204.32 420.120 < 2e-16 *** #> nitrogen:management 2 0.71 0.36 0.731 0.48341 #> nitrogen:variety 1 9.22 9.22 18.957 2.85e-05 *** #> management:variety 2 0.43 0.21 0.440 0.64531 #> nitrogen:management:variety 2 0.84 0.42 0.867 0.42278 #> Residuals 119 57.87 0.49 #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1