Example 8.3 from Generalized Linear Mixed Models: Modern Concepts, Methods and Applications by Walter W. Stroup (p-255)
Source:R/Exam8.3.R
Exam8.3.RdExam8.3 explains Response surface design with incomplete blocking
References
Stroup, W. W. (2012). Generalized Linear Mixed Models: Modern Concepts, Methods and Applications. CRC Press.
Author
Muhammad Yaseen (myaseen208@gmail.com)
Adeela Munawar (adeela.uaf@gmail.com)
Examples
## Response Surface Design with incomplete blocking (page 255)
data(DataSet8.3)
DataSet8.3$block <- factor(x = DataSet8.3$block)
DataSet8.3$aa <- factor(x = DataSet8.3$a)
DataSet8.3$bb <- factor(x = DataSet8.3$b)
DataSet8.3$cc <- factor(x = DataSet8.3$c)
library(lmerTest)
library(lattice)
Exam8.3.fm1 <-
lmer(
y ~ aa:bb:cc + a + b + c +
I(a^2) + I(b^2) + I(c^2) +
a*b + a*c + b*c + (1|block)
, data = DataSet8.3
)
#> fixed-effect model matrix is rank deficient so dropping 24 columns / coefficients
##--- page 256
anova(Exam8.3.fm1, ddf = "Kenward-Roger", type = 1)
#> Type I Analysis of Variance Table with Kenward-Roger's method
#> Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
#> a 40.960 40.960 1 12 19.1374 0.0009048 ***
#> b 45.901 45.901 1 12 21.4458 0.0005791 ***
#> c 4.306 4.306 1 12 2.0117 0.1815368
#> I(a^2) 22.103 22.103 1 3 10.3272 0.0488220 *
#> I(b^2) 35.975 35.975 1 3 16.8084 0.0262537 *
#> I(c^2) 1.495 1.495 1 3 0.6984 0.4646625
#> a:b 1.620 1.620 1 12 0.7569 0.4013689
#> a:c 2.205 2.205 1 12 1.0302 0.3301347
#> b:c 10.811 10.811 1 12 5.0512 0.0441960 *
#> aa:bb:cc 1.441 0.480 3 12 0.2245 0.8775621
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Exam8.3.fm2 <-
lmer(
y ~ a + b + c +
I(a^2) + I(b^2) + I(c^2) +
a*b + a*c + b*c + (1|block)
, data = DataSet8.3
)
##--- page 257
anova(Exam8.3.fm2, ddf = "Kenward-Roger", type = 1)
#> Type I Analysis of Variance Table with Kenward-Roger's method
#> Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
#> a 40.960 40.960 1 15 22.6507 0.0002534 ***
#> b 45.901 45.901 1 15 25.3828 0.0001471 ***
#> c 4.306 4.306 1 15 2.3810 0.1436508
#> I(a^2) 18.675 18.675 1 3 10.3272 0.0488220 *
#> I(b^2) 30.395 30.395 1 3 16.8084 0.0262537 *
#> I(c^2) 1.263 1.263 1 3 0.6984 0.4646625
#> a:b 1.620 1.620 1 15 0.8959 0.3588953
#> a:c 2.205 2.205 1 15 1.2194 0.2868867
#> b:c 10.811 10.811 1 15 5.9786 0.0273022 *
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
##--- page 257
Exam8.3.fm3 <-
lmer(
y ~ a + b + c +
I(a^2) + I(b^2) +
a*c + b*c + (1|block)
, DataSet8.3
)
anova(Exam8.3.fm3, ddf = "Kenward-Roger", type = 1)
#> Type I Analysis of Variance Table with Kenward-Roger's method
#> Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
#> a 40.960 40.960 1 16 22.7991 0.0002067 ***
#> b 45.901 45.901 1 16 25.5491 0.0001172 ***
#> c 4.306 4.306 1 16 2.3966 0.1411506
#> I(a^2) 20.067 20.067 1 4 11.1695 0.0287800 *
#> I(b^2) 32.660 32.660 1 4 18.1793 0.0130150 *
#> a:c 2.205 2.205 1 16 1.2273 0.2843011
#> b:c 10.811 10.811 1 16 6.0177 0.0260104 *
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
##--- scatter plot with regression plane by using Hoerl function ( page#233)
a <- seq(from = -1, to = 1, by = 1)
b <- seq(from = -1, to = 1, by = 1)
c <- seq(from = -1, to = 1, by = 1)
abc <- expand.grid(a = a, b = b, c = c)
Yhat <- NULL
for(i in 1:nrow(abc)) {
Yhat[i] <- 50.08500 + 1.6*abc$a[i] + 1.69375*abc$b[i] + 0.51875*abc$c[i]-
3.30250*I((abc$a[i])^2)-3.51500*I((abc$b)^2)[i] -
0.52500*(abc$a)[i]*(abc$c)[i]-1.16250*(abc$b)[i]*(abc$c)[i]
}
Newdata <- data.frame(abc, Yhat)
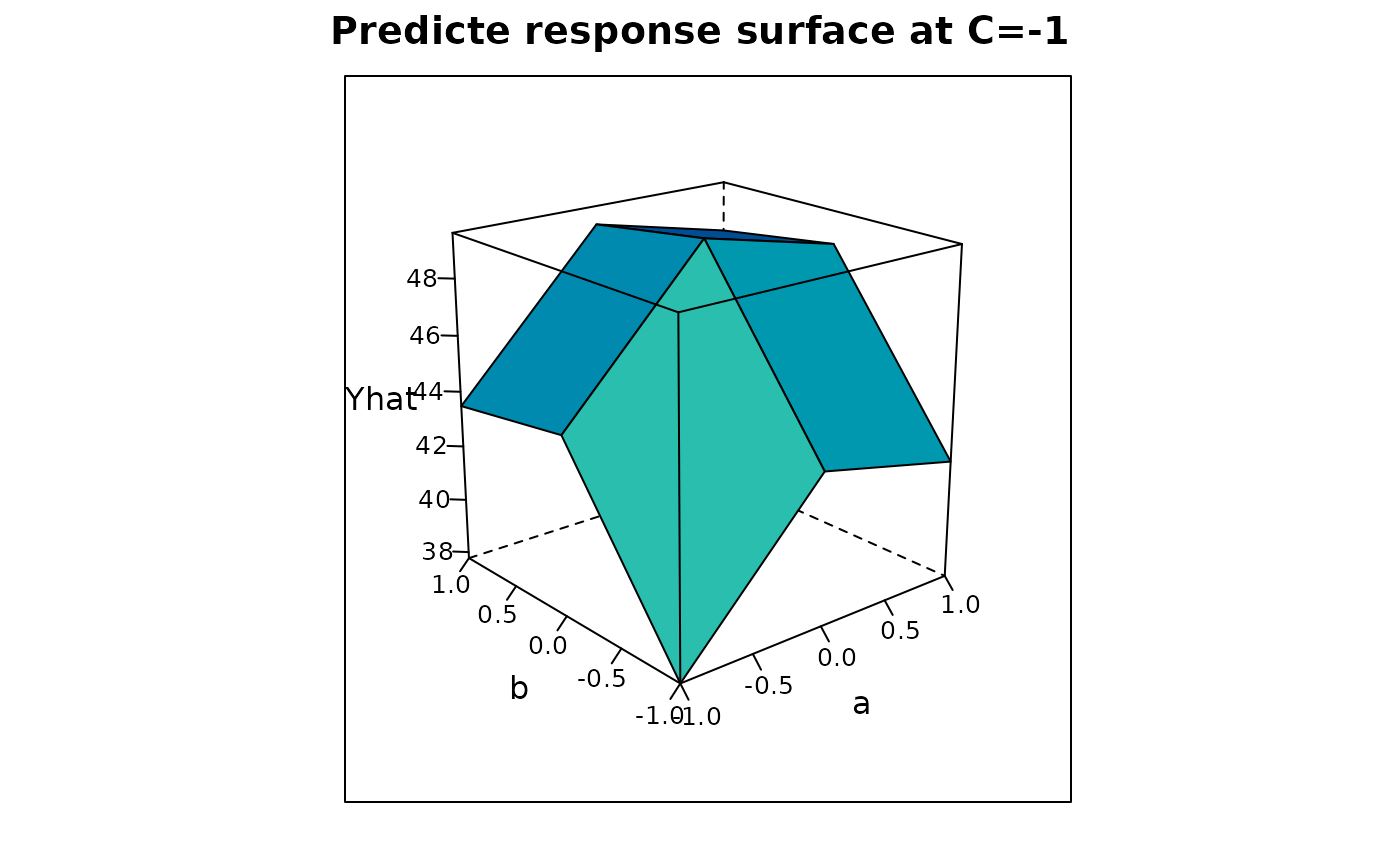
Plot1 <-
wireframe(Yhat ~ b*a, data = subset(Newdata,c==-1),
xlab = "b", ylab = "a",
main = "Predicte response surface at C=-1", colorkey = FALSE,
drape = TRUE, scales = list(arrows = FALSE),xlim=c(max(b),(min(b))),
screen = list(z = -50, x =-70)
)
Plot2 <-
wireframe(Yhat ~ b*a, data = subset(Newdata,c==0),
xlab = "b", ylab = "a",
main = "Predicte response surface at C=0", colorkey = FALSE ,
drape = TRUE, scales = list(arrows = FALSE),xlim=c(max(b),(min(b))),
screen = list(z = -50, x =-70)
)
Plot3 <-
wireframe(Yhat ~ b*a, data = subset(Newdata,c==1),
xlab = "b", ylab = "a",
main = "Predicte response surface at C=1", colorkey = FALSE,
drape = TRUE, scales = list(arrows = FALSE),xlim=c(max(b),(min(b))),
screen = list(z = -50, x =-70)
)
print(Plot1)
 print(Plot2)
print(Plot2)
 print(Plot3)
print(Plot3)
